Introduction to bioinformatics
What is Bioinformatics
Bioinformatics is the study of biology through the efficient analysis of large datasets. What is a large dataset? One rule of thumb is that a dataset is large if one of its components has more than 2^{16} = 65,536 pieces; many data sets are much larger, with the raw data taking up petabytes (10^{15}) or more of storage. It is interdisciplinary, involving biology, computer science, statistics, mathematics, and engineering. It is a large and rapidly growing subject with many aspects.
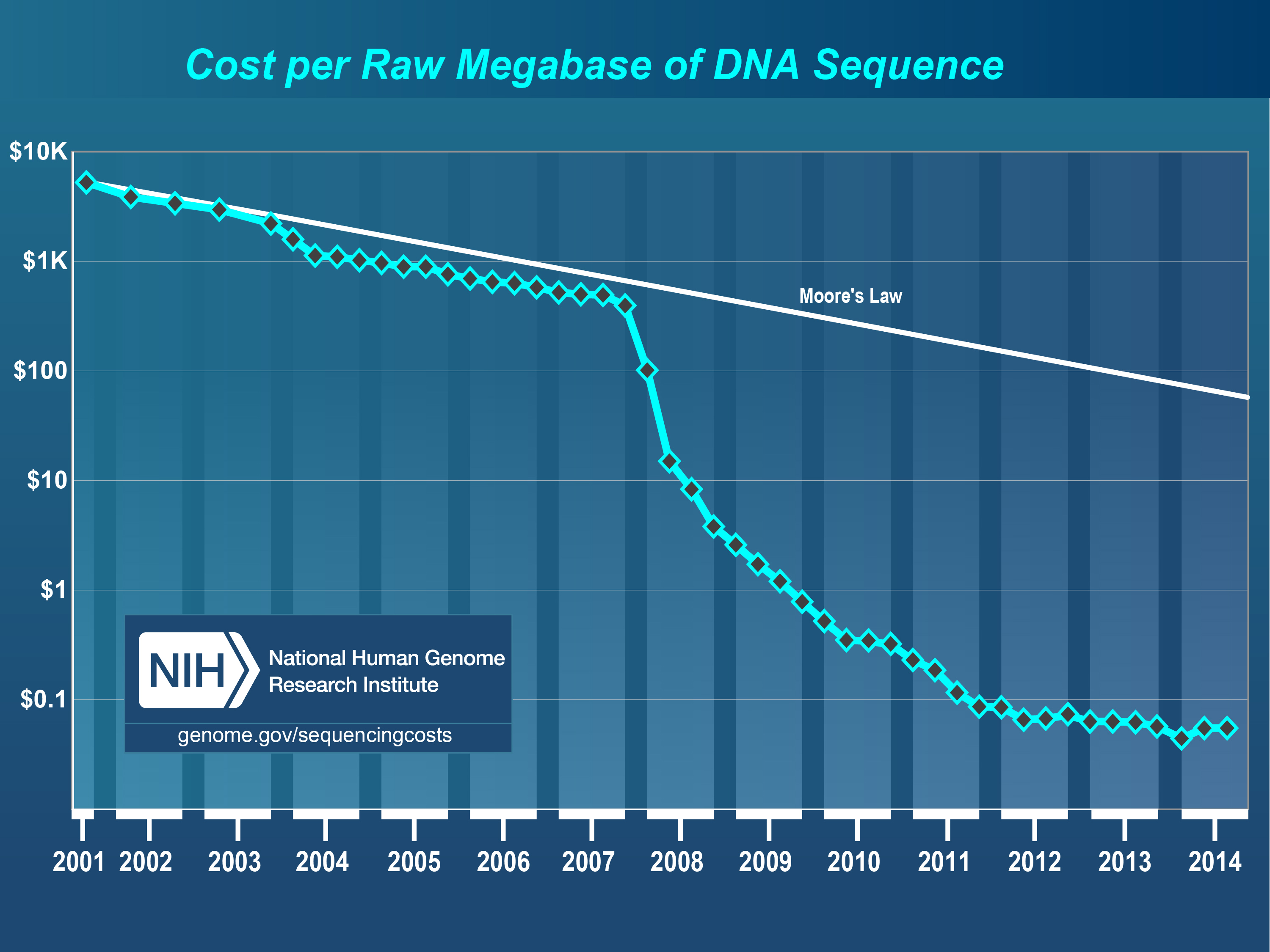
Figure 1.1 shows a remarkable event in the history of human technology. For the last 70 years the density of transistors on computer chips has roughly doubled every two years, a phenomenon sometimes referred to as Moore's Law (named after a former CEO of the Intel Corporation). This amazing sustained growth has resulted in the ubiquity of powerful computing devices including smartphones. In the figure, which is plotted on a semilog scale, a decrease in cost corresponding to Moore's Law appears as a straight line. We can see that from 2001 to 2007 improvements in DNA sequencing resulted in lower costs at a rate similar to Moore's Law. This was an impressive achievement and scientists had to deal with rapidly expanding data sets. Then around 2007 we see a truly astounding super-exponential drop in cost, which massively outpaced the improvements in computing power. This has resulted in a staggering production of data from DNA and RNA.
A hallmark of bioinformatics is its iterative dependence on data. An example of this is the use of protein sequence alignments to construct empirical amino acid scoring matrices; the scoring matrices are then used to align new and more distantly related protein sequences. This spiral (not circular!) of data-driven development is one of the primary characteristics of an informatics discipline.
The vast amount of DNA sequencing that has been done is concentrated on a relatively small number of species. As of October 2018, the twenty most sequenced organisms (as reported by NCBI's GenBank) are listed in the table below. The number of bases reported is for the final assembled and processed data; the amount of raw sequence behind these is several orders of magnitude larger. It can be helpful to be familiar with these model organisms, as much of the data from other species is analyzed based on them. Listing by total bases is somewhat misleading, since it is biased towards organisms with large genomes such as wheat and barley. The primary model organism among plants is Arabidopsis thaliana, which was chosen partially because of its small genome. Despite the economic importance of plants such as wheat and barley, there is far more experimental genetic data about A. thaliana, even though it does not make it on this list.
| Entries | Bases | Species | Common Name |
|---|---|---|---|
| 26631300 | 19772380478 | Homo sapiens | Human |
| 1936389 | 17184977305 | Triticum aestivum | Wheat |
| 9899387 | 10247302143 | Mus musculus | Mouse |
| 2199524 | 6530109968 | Rattus norvegicus | Rat |
| 2232175 | 5431743022 | Bos taurus | Cow |
| 4210879 | 5248587993 | Zea mays | Corn |
| 3301603 | 5076267171 | Sus scrofa | Pig |
| 126307 | 3302097453 | Escherichia coli | E. coli |
| 1346898 | 3237283130 | Hordeum vulgare | Barley |
| 1729679 | 3191499108 | Danio rerio | Zebrafish |
| 1239296 | 2836942615 | Oryzias latipes | Medaka (rice fish) |
| 311591 | 2682429646 | Arachis hypogaea | Peanut |
| 72 | 2590574434 | Ovis canadensis canadensis | Bighorn sheep |
| 746614 | 2572331056 | Solanum lycopersicum | Tomato |
| 111 | 2290221631 | Bos mutus | Yak |
| 205550 | 1836754082 | Cyprinus carpio | Carp |
| 1379151 | 1727234169 | Oryza sativa | Rice |
| 327068 | 1595510956 | Apteryx australis mantelli | Brown kiwi |
| 1657 | 1485366977 | Bordetella pertussis | causes whooping cough |
| 12860 | 1468683875 | Klebsiella pneumoniae | causes pneumoniae |
In this text we will concentrate on three main topics: primary sequence analysis, phylogenetics, and machine learning.
Recommended reading
This text draws on many sources, not all of which are listed below. It provides only an introduction into each topic. The following references are highly recommended if you wish to obtain a deeper understanding into one of the areas they cover.
-
Coalescent theory, by John Wakeley. A very readable introduction into this field, which complements the more commonly taught classical population genetics.
-
The Origins of Genome Architecture, by Michael Lynch. The primary theme of this book is the application of population genetics ideas on selection to the evolution of genes and genomes.
-
Molecular Evolution: A Statistical Approach, by Ziheng Yang. An excellent text on sequence evolution and phylogenetics.
-
Statistical Methods in Bioinformatics, by Warren J. Ewens and Gregory R. Grant. This book focuses on the statistical foundations of sequence evolution, sequence alignment and scoring, phylogenetics, and hidden Markov models.
-
Inferring Phylogenies, by Joe Felsenstein. This is a fantastic reference for all phylogenetic methods developed up to its publication in 2004, by one of the pioneers of the subject.
-
Biochemical pathways, edited by Gerhard Michal and Dietmar Schomburg. This book is not on bioinformatics; it provides a high level overview of the most important biochemical pathways in all living creatures. This knowledge provides important context for interpreting bioinformatic data.
-
Biological Sequence Analysis: Probabilistic Models of Proteins and Nucleic Acids, by Richard Durbin, Sean R. Eddy, Anders Krogh, and Graeme Mitchison. A definitive text for bioinformatics that provides deep insight into the use of hidden Markov models and stochastic context-free grammars in sequence and structure analysis.
-
Bioinformatics and Molecular Evolution, by Paul G. Higgs and Terri Attwood. This is a little out of date in places but its presentation heavily influenced many of the topics herein.
-
Information Theory, Inference, and Learning Algorithms, by David J.C. MacKay (this has a free online PDF). A beautifully clear book on Bayesian learning and information theory.
-
Modern Statistics for Modern Biology, by Susan Holmes and Wolfgang Huber. You can buy a physical version of this text but the online form is very well done.
Computing
In this text we will use the programming language Python, along with the extension module Biopython. Python is a high-level language developed since 1991, and is very widely used in bioinformatics. Other popular high-level languages for bioinformatics include Perl, Ruby, Java, and R. For applications requiring efficient and speedy computation, C, C++, and Java are common choices, but in many workflows these specialized applications are used within a high-level scripting environment such as Python.
There are many excellent resources online for learning Python. A particularly relevant one for bioinformatics is the Rosalind site, which has a great collection of practice problems. Another convenient site with an interactive Python tutorial is hosted here. For a general introduction to Python in book form, the free Think Python by Allen Downey is quite good.
There are a myriad of helpful online services for bioinformatics; below are links to some of the most generally useful for the material in this text:
Essentials of molecular biology
It is beyond the scope of this book to give an adequate introduction to molecular biology, and if you are unfamiliar with this subject we strongly urge you to learn more about it from texts, videos, and other courses.
Biological scale
It can be helpful to have some understanding of the approximate size of various entities in biological systems. There is an excellent text, Cell Biology by the Numbers, which provides measurements and estimates for a wide variety of biological entities and processes. This has an associated database at the website http://book.bionumbers.org/, which is very helpful for quantitative modeling and estimation. Here we provide a small sample of the most important extensive quantities.
DNA base pair length \approx 0.33 \ \text{nm}.
Thickness of lipid bilayer: 5 \ \text{nm}.
Nucleosome diameter: 10 \ \text{nm}.
Volume of typical protein: 25 \ \text{nm}^3.
Mass of typical amino acid: 100 Daltons.
E. coli cell volume \approx 1 to 2 \ \mu \text{m}^3.
Yeast cell volume \approx 60 \ \mu \text{m}^3
Longest human neurons: 1 \ \text{m}.
DNA
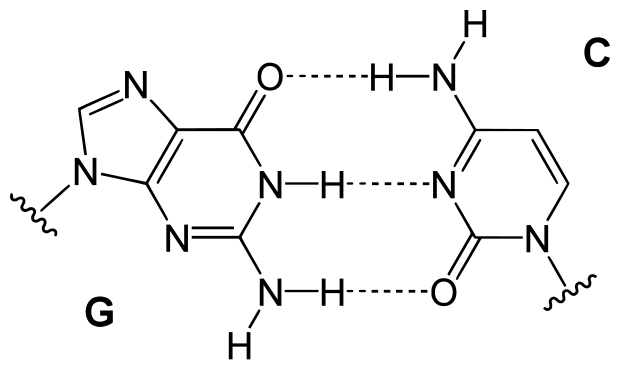
Deoxyribonucleic acid, or DNA, is a long stable polymer composed of nucleotide units (adenine (A), cytosine (C), guanine (G), and thymine (T)) linked together by phosphate-sugar units (the deoxyribose). In cells the DNA is usually in a two-stranded form in which the two strands are linked with many hydrogen (non-covalent) bonds between the nucleotide units.
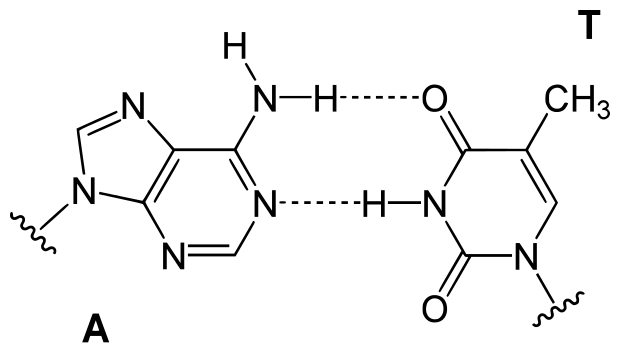
The strands will bond most stably when the nucleotides from the two strands pair up in Crick-Watson pairs: A-T and C-G. It is important to note that these two kinds of pairs are not energetically the same, since the C-G pairs form three bonds while the A-T pairs form only two.
RNA
Ribonucleic acid, or RNA, is very similar to DNA but the differences are crucially important for its biological role. The first difference is that RNA usually uses the nucleotide uracil (U) instead of thymine. The second difference, that the phosphate-sugar bakcbone uses ribose instead of dexyribose, is more important. The extra hydroxyl (-OH) group in the ribose allows many chemical reactions that are not possible for DNA, including the possibility of the phosphate group to cleave the backbone. This means that RNA is much more unstable than DNA, and it makes sense that cells use DNA for information storage, while RNA is used for short-term message transmission as well as many other enzymatic roles. However, some viruses use RNA instead of DNA to store their genome.
There are many kinds of RNA in the cell. In fact, the diversity of functions of RNA suggests the 'RNA World' hypothesis: that the earliest forms of life were RNA-based, with the use of DNA arising later. Although this hypothesis is difficult to test, there is a great deal of indirect evidence for it. We will briefly survey the most important kinds of RNA:
-
Messenger RNA (mRNA). This type of RNA is part of the classic 'Central Dogma' of molecular biology: that information is instantiated from nucleic acids into proteins, but not the reverse. In particular, protein-coding DNA is transcribed into mRNA, which is then translated into proteins by ribosomes. The ribosomes and the translation process involve two other kinds of RNA (described next).
-
Ribosomal RNA (rRNA). Ribosomes are complicated structures with two main subunits formed from many proteins and four RNA strands. The two ribosomal subunits are slightly different in prokaryotes (bacteria and archaea, which lack cell nuclei) and eukaryotes (all other life apart from viruses; this includes all plants, fungi, and animals). In eukaryotes the subunits are named 60S and 40S; the 'S' refers to a sedimentation rate in a centrifuge; in prokaryotes they are the 50S and 30S subunits. The larger subunit has three rRNA strands, and the smaller subunit has one. In humans, the longest of the rRNA pieces is the 28S, and is 5070 nucleotides. There are many ribosomes in a cell, which means that usually the rRNA is the most abundant type of RNA.
-
Transfer RNA (tRNA). Transfer RNA is the critical bridge between the genetic code in mRNA and the amino acid sequence of the translated protein. In every organism there are numerous tRNAs which vary in which codon and amino acids they will bind to.
-
Small nuclear and small nucleolar RNA (snRNA and snoRNA). As the names suggest, this type of RNA is usually only present in the nucleus, where it helps process and modify ribosomal and transfer RNA.
-
Small interfering RNA (siRNA). These short fragments (about 20 to 24 base pairs) can reduce the gene expression of complementary mRNA; synthetic fragments are often used to study the effects of perturbed gene expression.
In addition to the bases A,C,G,T, and U, there are single-letter abbreviations for combinations of bases as follows:
| Abbreviation | Meaning |
|---|---|
| A | Adenine |
| C | Cytosine |
| G | Guanine |
| T (or U) | Thymine (or Uracil) |
| R | A or G |
| Y | C or T |
| S | G or C |
| W | A or T |
| K | G or T |
| M | A or C |
| B | C or G or T |
| D | A or G or T |
| H | A or C or T |
| V | A or C or G |
| N | any base |
| . or - | gap |
NOTE: because RNA, which has U instead of T, is often sequenced in the form of DNA it is very common to use the letter T to denote the RNA base uracil in databases.
Proteins
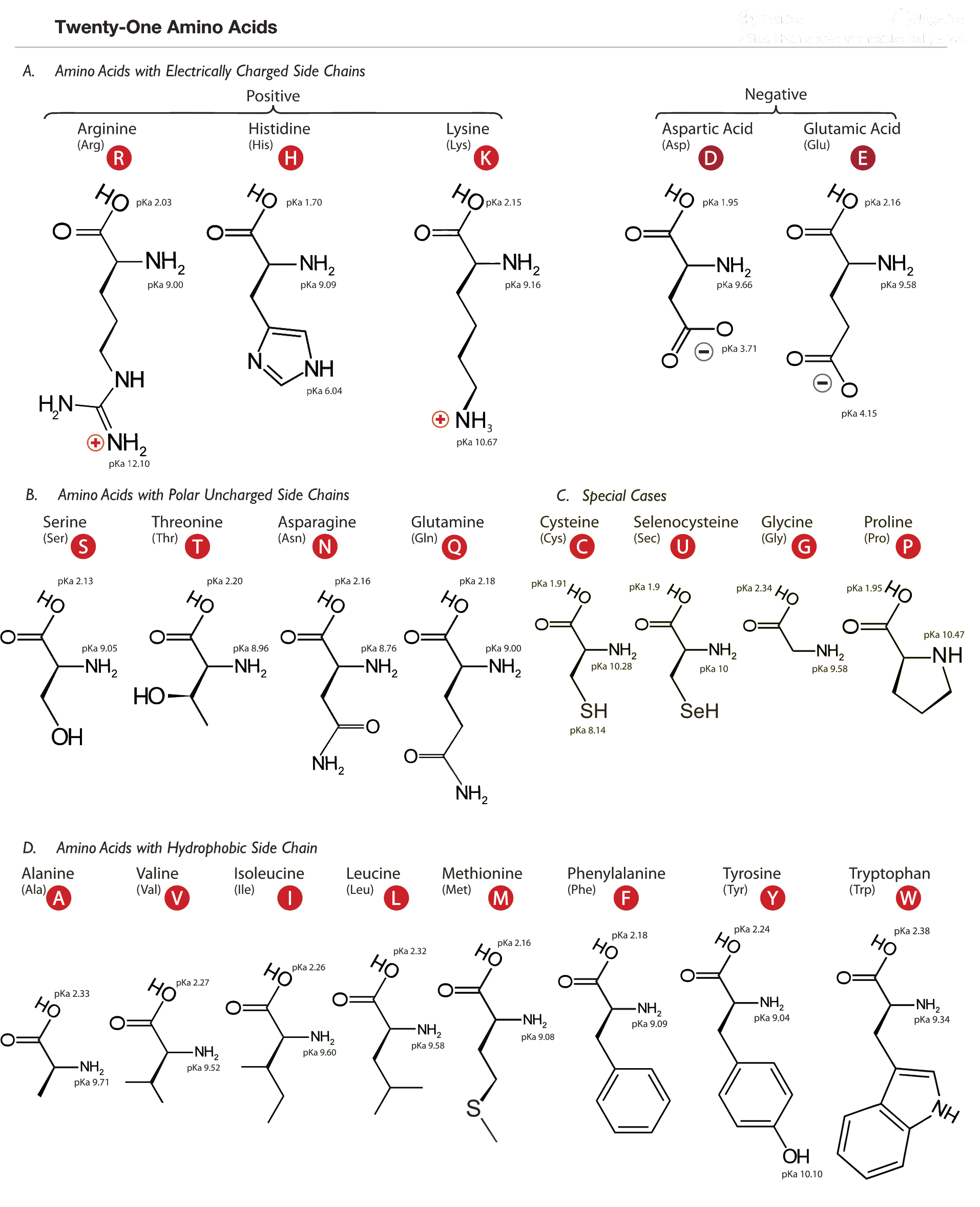
Proteins are linear polymers formed from amino acid units, joined by carbon-nitrogen peptide bonds. Although the backbone always consists of carbon-carbon-nitrogen triplets, there are many kinds of amino acids because of their varying sidechains. The twenty amino acids that make up standard proteins are shown below. Other amino acids can be involved in cellular processes but are not incorporated into proteins, such as citrulline. There are also some amino acids that are modified after or during the translation process; one of them, selenocysteine, is shown in the chart below. Selenocysteine is modified from cysteine by replacing the sulfur atom with a selenium.
If you plan on doing any sort of work with protein information, it is crucial to get some sense of the chemical and physical differences between the amino acids. The wide variety of sidechains is what gives proteins their tremendous range of chemical abilities and physical properties. The chart below categorizes them by one of their most important aspects, the charge and hydrophobicity of the side chains. Asparagine (N) and glutamine (Q), while listed as uncharged, are highly polar and more hydrophilic than some of the charged amino acids. Cysteine can be critically important in proteins because of its ability to form disulphide bonds. Proline is unique in that its sidechain loops around and covalently bonds the protein backbone again, which reduces the flexibility of the peptide bond at that point.
Triplets of DNA bases - codons - code for amino acids in proteins through an almost universal code. Some codons are different in different organisms, but only in few places. For example, the codon TGA in humans (and many other eukaryotes) codes for tryptophan instead of being a stop codon. Here we adopt the convention of most sequence databases of using T instead of U, despite the U being in the messenger RNA.
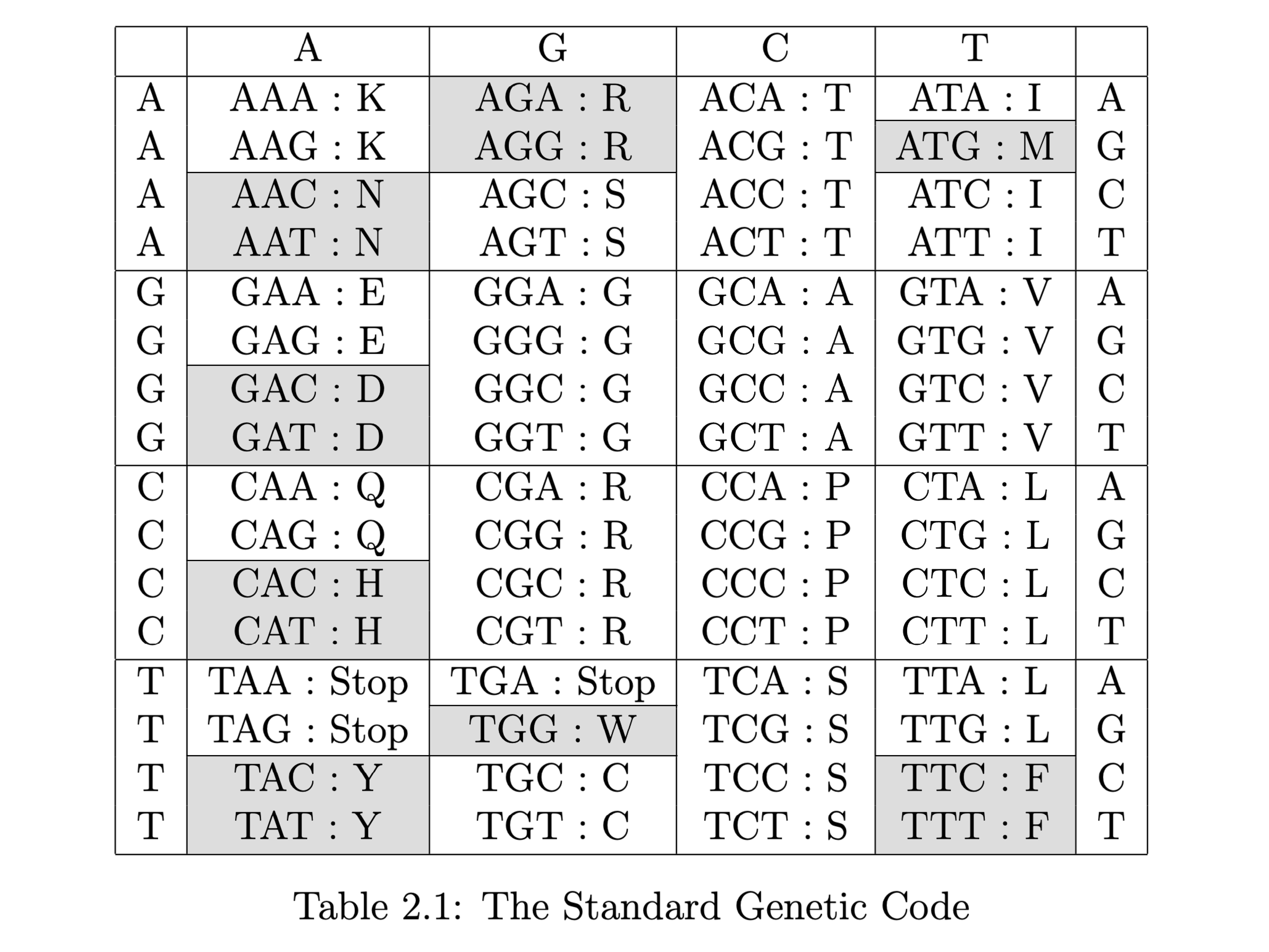 The Standard Genetic Code
The Standard Genetic Code
Of the many slightly different versions of the genetic code, those used by mitochondria are among the most different. One of the most important differences between mitochondrial genetic codes and the standard code is that the codon TGA encodes for tryptophan (W) rather than being a stop codon.
The cell
We will briefly survey some of the structure of typical cells. The reader is strongly encouraged to learn about cells in more depth from more focused literature.
Cell membrane
The cell membrane defines the cell itself. In Bacteria and Eukaryota it mostly consists of a double layer of lipids, along with membrane-bound and spanning proteins. In the Archaeal branch of life, the lipid membrane is a double-headed monolayer of isoprenoid lipids. The membrane proteins perform many different functions, most notably signalling and transport. For example, the interior of most cells has a low concentration of sodium and chloride ions, with a high concentration of potassium ions. These differences in ion concentration must be actively maintained by protein pumps on the cell membrane. The difference in ion concentrations also usually results in a voltage across the cell membrane, which can be used to drive chemical reactions.
An important example of a class of membrane proteins is the G protein-coupled receptor family, also known as seven-transmembrane domain receptors. Most of the members of this family perform signal transduction functions.
Mitochondria and Chloroplasts
Mitochondria are important elements of a cell for many reasons. Perhaps the most important is that their inner membrane contains protein complexes which use various substrates (usually derived from glucose) to produce an electrochemical gradient across the membrane. This gradient is used to power the ATP synthase complex, which produces ATP (adenosine triphosphate). ATP is used universally by all life as we know it to drive energetically unfavorably chemical reactions. Other mitochondrial complexes are important in calcium and iron homeostasis and lipid metabolism.
The leading theory on the origin of mitochondria is the endosymbiotic hypothesis - that mitochondria were originally bacteria which were somehow incorporated into another cell, about two billion years ago. Over time, much of the mitochondrial DNA shifted to the nucleus. In mammals, only 13 protein-coding genes are present in the mitochondrial genome. Hundreds of nuclear-encoded proteins are transferred to the mitochondria after being translated.
Chloroplasts are similar in that they are endosymbiotically derived organelles, but their thylakoid membranes contain complexes involving the molecule chlorophyll to transduce light into high energy bonds while producing oxygen from water (photosynthesis). Chloroplasts, like mitochondria, contain their own small genome.
Nucleus
Not all cells have nuclei; those that do are called eukaryotes, and those that do not are called prokaryotes. The prokaryotes consist of two large domains, the Bacteria and the Archaea. The eukaryotes have many subdomains, including all known multicellular life (plants, animals, fungi, etc).
The nucleus has a double-layer lipid membrane just like the cell itself. Inside the nucleus is all of cell's DNA, with the exception of the DNA of the mitochondria and chloroplasts.
Endoplasmic reticulum
Eukaryotes have an extensive membrane system attached to the nucleus called the endoplasmic reticulum (ER). This organelle performs many functions including protein synthesis, lipid metabolism, and protein transport. The endoplasmic reticulum often has contacts and connections with the mitochondrial membranes.
Golgi apparatus
Another membrane system found in most cells is the Golgi apparatus, which is usually located near the endoplasmic reticulum. Proteins that are translated in the ER are sometimes shipped to the Golgi apparatus in lipid vesicles, and then further processed. Some of these proteins are then exported out of the cell, again in vesicles, or incorporated into the cell membrane.
Cytoskeleton
An extremely important component of every living cell is the cytoskeleton, a network of filamentary proteins. Besides providing mechanical properties to the cell, it is used for protein transport, locomotion, and in cell division.
The Domains of Life
As mentioned above in the discussions of cell membranes and the nucleus, on the highest level it makes sense to classify life into three main branches: the Archaea, the Bacteria (or Eubacteria), and the Eukarya (or Eukaryota). The split between the Archaea and Bacteria was proposed in 1977 by Woese and Fox, based on comparisons of ribosomal RNA. Before that analysis the anucleate Archaea and Bacteria were lumped together as 'prokaryotes'. The Eukaryotes are thought to have arisen from an ancient combination of an archaeal host and an endosymbiotic bacterium.
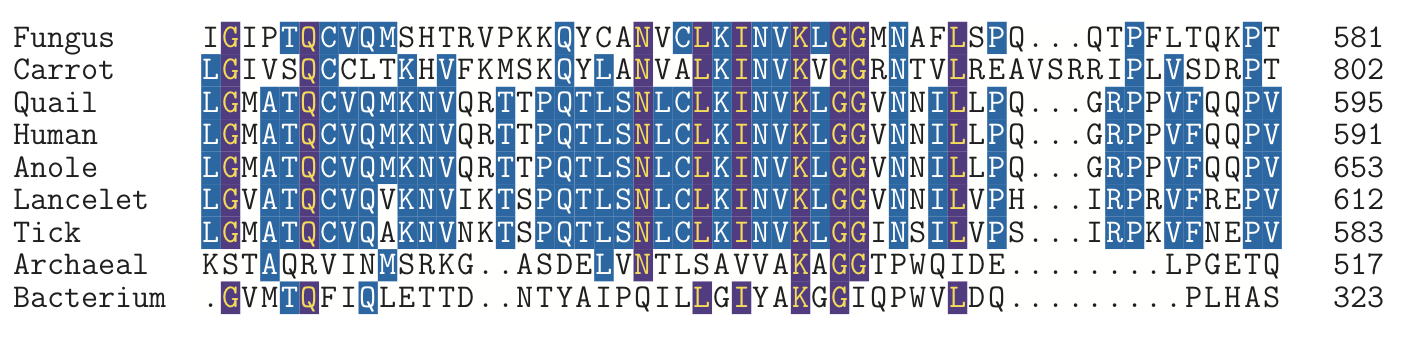
It is remarkable that all of life shares a very similar genetic code. There are small differences between some lineages in the code, but it seems clear that the fundamental process of translation has descended without major changes from a common ancestor at least 3.5 billion years ago. Because of the enormous time that has elapsed between the three lineages of life, it is usually difficult to compare DNA or protein sequences between them even when the function of those sequences has remained similar. The main exceptions are ribosomal and transfer RNAs, and some highly conserved proteins such as the nucleotide binding PIWI and Argonaut family. Below we show a small portion of a multiple protein sequence alignment from a variety of Eukaryotes and single representatives from the Archaea and Bacteria: