Understanding the details of protein function is a fundamental step in developing new therapies to treat disease. In our lab, we develop and employ molecular modelling techniques to study the structure, dynamics and function of proteins involved in clinical disorders, and to characterize the physicochemical properties of small organic molecules. The two broad research questions that we are currently aiming to answer are: (1) what are the determinants that define allostery in proteins; and (2) how does the membrane environment affect the function of proteins? To address these questions we study how a few selected proteins carry out their function.
Allosteric Response in Calcium Binding Proteins
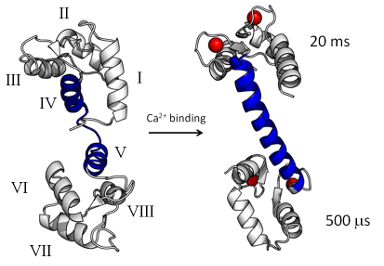 As a paradigm for allosteric response in proteins we study calmodulin (CaM), a small ubiquitous
calcium-sensing messenger that binds and regulates over 300 different target proteins and plays a crucial
role in numerous cellular processes. X-ray crystallography, IR, and NMR have identified conformational
dynamics as key to understanding CaM function, yet an atomistic explanation of the response mechanism and
adaptation of CaM to its multiple targets is incomplete. It has also been shown that oxidation of
methionine residues in CaM impairs formation of the complex between CaM and some of its targets. We employ
computer simulations to focus on three aims: (1) explain the modulation of CaM activity in terms of the
specific interactions between CaM, its targets, and the solvent; (2) understand the degree of cooperativity
and the mechanism at the base of CaM binding affinity and adaptation to its numerous targets; and (3)
identify the specific interactions between CaM and its targets that are altered upon CaM oxidation.
As a paradigm for allosteric response in proteins we study calmodulin (CaM), a small ubiquitous
calcium-sensing messenger that binds and regulates over 300 different target proteins and plays a crucial
role in numerous cellular processes. X-ray crystallography, IR, and NMR have identified conformational
dynamics as key to understanding CaM function, yet an atomistic explanation of the response mechanism and
adaptation of CaM to its multiple targets is incomplete. It has also been shown that oxidation of
methionine residues in CaM impairs formation of the complex between CaM and some of its targets. We employ
computer simulations to focus on three aims: (1) explain the modulation of CaM activity in terms of the
specific interactions between CaM, its targets, and the solvent; (2) understand the degree of cooperativity
and the mechanism at the base of CaM binding affinity and adaptation to its numerous targets; and (3)
identify the specific interactions between CaM and its targets that are altered upon CaM oxidation.
Membrane Proteins
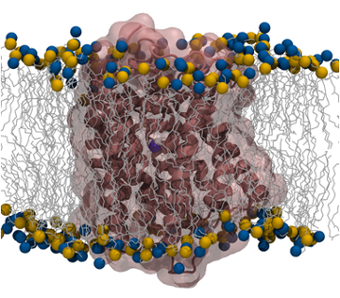 To probe the interplay between proteins and biological membranes, we investigate a variety of membrane
proteins that are characterized by different topology and function. We study small molecule transporters
such as the ammonia transporter, AmtB, to understand how permeation through the membrane of such molecules
can be facilitated. To understand how membrane proteins re-shape and alter membrane properties, we study the
mechanism of insertion and aggregation of antimicrobial peptides in lipid bilayers. We are also interested
in probing the subtle effects that characterize the interactions at the water/lipid interface by studying
how protein unfolded domains can be ordered upon membrane interactions.
To probe the interplay between proteins and biological membranes, we investigate a variety of membrane
proteins that are characterized by different topology and function. We study small molecule transporters
such as the ammonia transporter, AmtB, to understand how permeation through the membrane of such molecules
can be facilitated. To understand how membrane proteins re-shape and alter membrane properties, we study the
mechanism of insertion and aggregation of antimicrobial peptides in lipid bilayers. We are also interested
in probing the subtle effects that characterize the interactions at the water/lipid interface by studying
how protein unfolded domains can be ordered upon membrane interactions.
Methods
We tackle these problems by using a variety of computational techniques, which include quantum mechanics (QM), molecular mechanics (MM), and their combined approach (QM/MM). We extend the reach of classical molecular dynamics (MD) simulations with enhanced sampling methods, and improve their accuracy by refining the underlying force fields to better reproduce experimental observables.