|
Berry Lab Research Projects |
|||
We are interested in the roles of metals in biological systems and particularly in the roles of copper in enzyme structure and function. We are also interested in the ways that proteins, through both primary and secondary coordination spheres, control the copper ion and tune its properties. We are always looking for hardworking, self-motivated students at both the undergraduate and MS levels to work with us on these and related projects. We provide all the necessary training in these multidisciplinary projects that can include such techniqes as PCR, DNA purification, recombinant protein expression and purification, enzyme assaying, electrochemistry, EPR, mass spectrometry, and crystallography. Please contact Dr. Berry for information on available lab positions. |
||||
|
||||
Modeling Copper Proteins |
||||
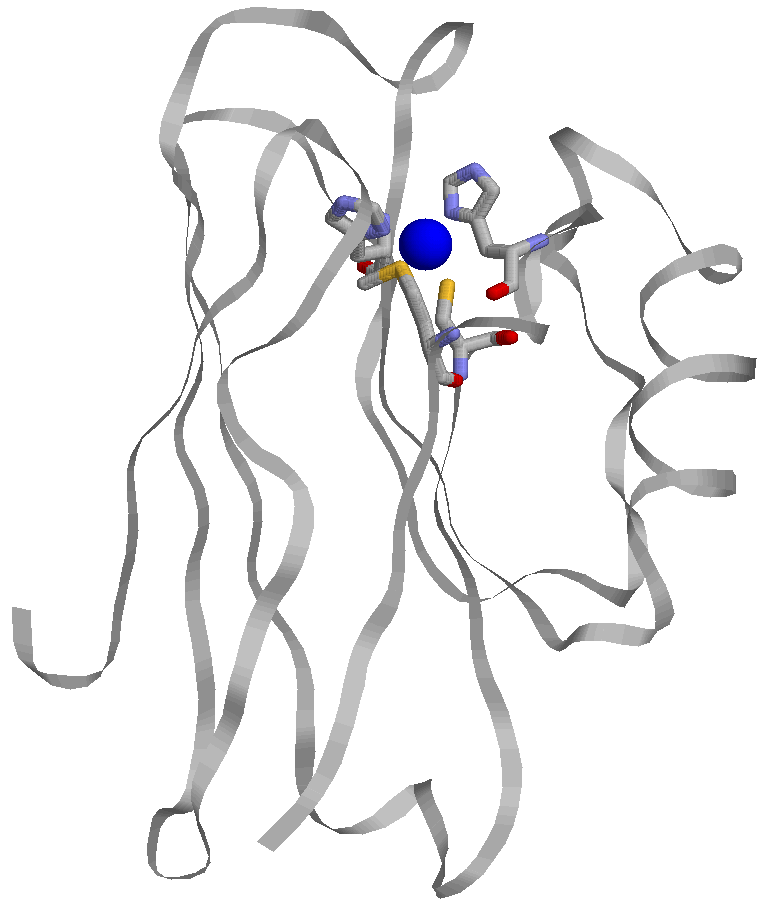
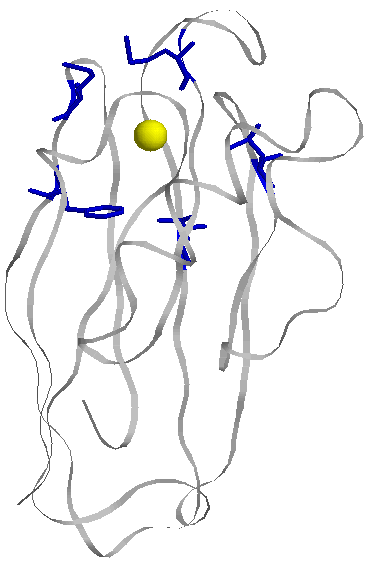
It is estimated that at least a third and as many as a half of all proteins utilize metal ions for proper function. Of these, copper proteins are an important class of metallobiomolecules. There are at least a dozen enzymes in the human body that utilize copper for structure and function, as well as many recently discovered Cu chaperone proteins. Common roles of many of these copper enzymes are electron transfer and reactivity with O2 and reduced species of O2 . Most human copper enzymes are classified as oxidoreductases and are involved in an array of processes. Amongst copper enzymes are the recurring themes of copper mediated electron transfer and copper catalyzed oxygen chemistry with both oxidation and reduction of substrate. The goal of our work is to use protein design and redesign to explore and elucidate these commonly occurring themes in copper metalloenzymes. We utilize an existing protein, azurin (Figure right), as the ligand or scaffold in which to design our models. |
 |
|||
|
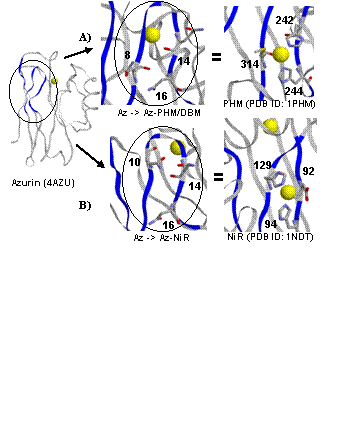 |
We are interested in the function of the 'noncoupled' copper proteins which contain two copper ions separated by over 10 angstroms. Examples of these proteins include the mammalian enzymes peptidylglycine alpha-hydroxylating monooxygenase (PHM), and dopamine beta-monooxygenase (DBM), and the bacterial nitirite reductases (NiR). We designed the catalytic, T2 Cu sites of PHM, DBM, and NiR onto the surface of azurin by mutating three residues in azurin which were arranged in a similar structural motif as those found in the active site of the target enzymes (Figure left). Importantly, this location maintains a similar separation (11-13 Å) from the existing T1 electron transfer site in azurin and thus closely mimics the arrangement of the noncoupled copper centers found in PHM, DBM, and NiR. The EPR spectra of these models resemble the native systems. Our group continues to study the spectroscopy, structure, and enzymatic activity of these models. We have so far identified low level NiR activity in these models. We have also grown X-ray diffration quality crystals of the PHM-Az variant and we are working to determine the structure (Figure right). |
 |
||
|
|||||
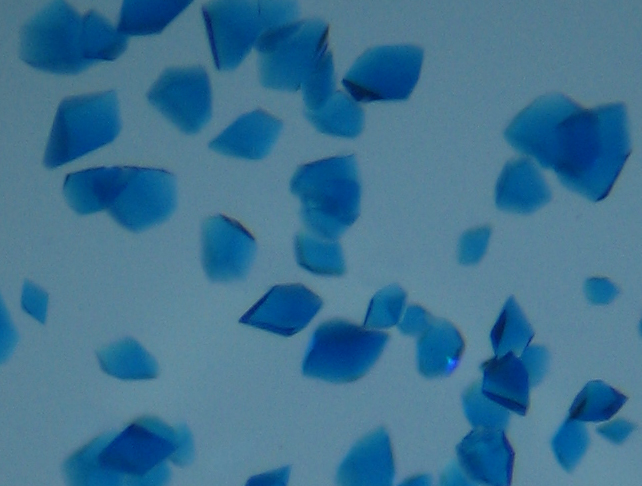
Fig. PHM-Az crystals (top) with and (bottom) without extra copper added. |
|||||
|
|||||
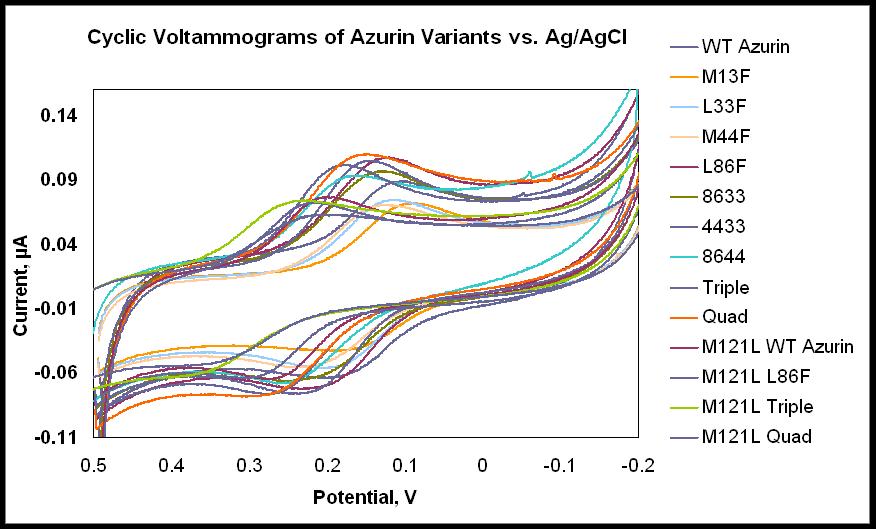 |
Our group is also interested in the effects of the secondary coordination sphere on copper site properties, such as reduction potential. We have made a number of mutations in azurin attempting to introduce hydrophobicity into the environment surrounding the copper ion. Figure (right) shows the residues we mutated to phenylalanine residues. We have measured modest increases in reduction potential with these mutants and we continue to explore the effects of such mutations on the reduction potential. |
 |
|||
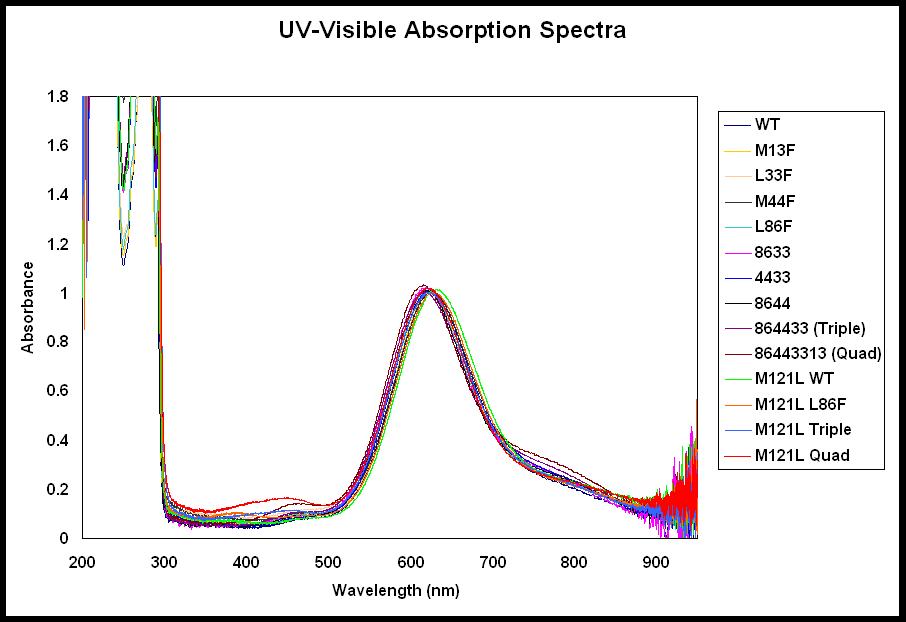 |
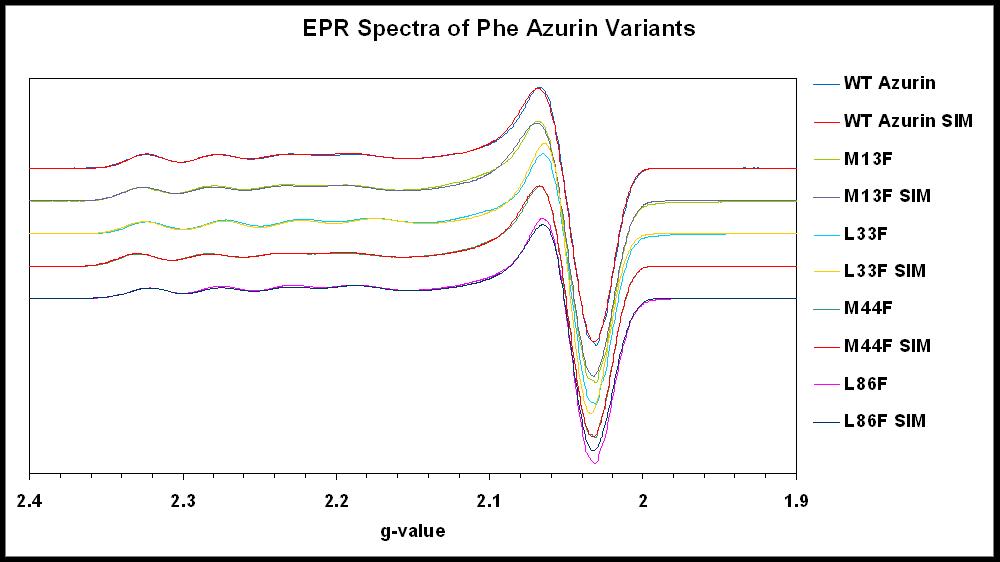
Our group is capable of characterizing our copper protein variants by cyclic voltammetry and UV-Visible absorption and EPR spectroscopy (left and below). |
||||
 |
|||||
|
Lanthanide coordination chemistry |
|||||
Our group has synthesized some unique lanthanide complexes including a series of dinuclear Eu(III), Sm(III), and Yb(III) beta-diketonate complexes with a bridging 2,2'-byprimidine ligand (Figure top right). These complexes are more examples amongst a small group of related dinuclear lanthanide species.
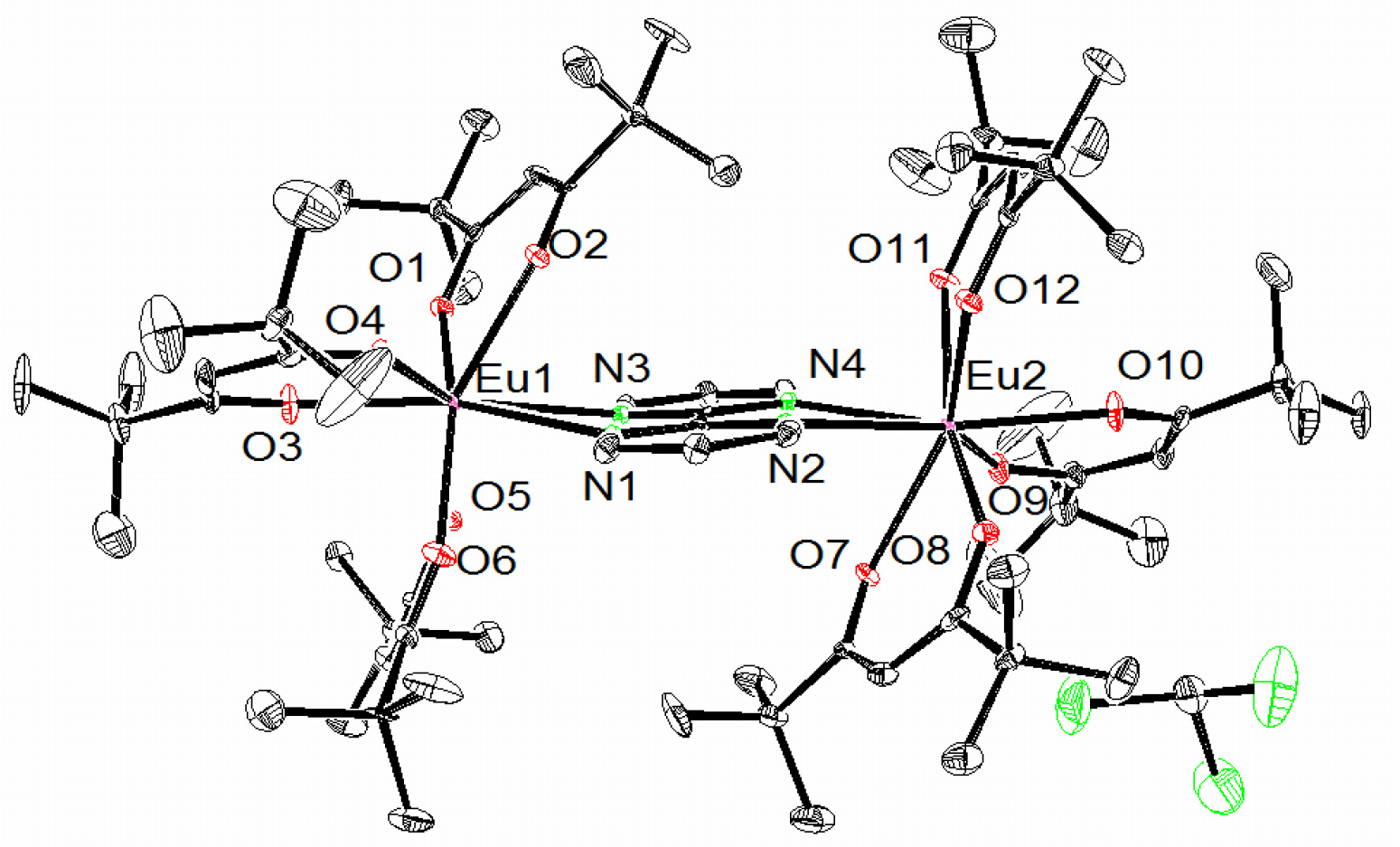
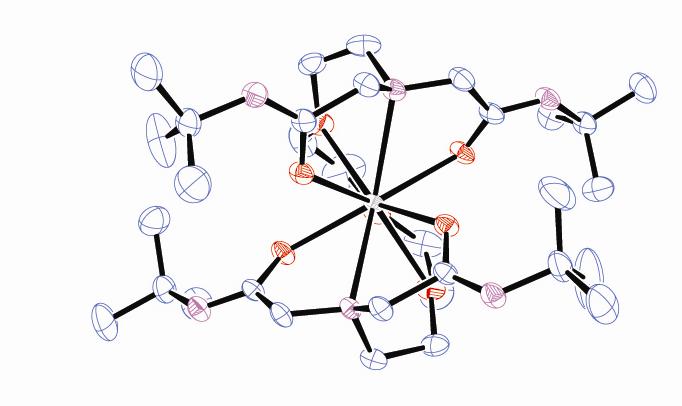
Our group has also synthesized a unique multi-dentate amide containing ligand that binds Ln(III) ions stably in solution. We were able to characterize the complexes by X-ray crystallography (Figure bottom right) and extensive 1- and 2-D NMR techniques. Finally, we demonstrated that our complexes displayed some PARACEST abilities, a new technique with possible MR imaging applications (Figure far right). |
 |
 |
||||
 |
||||||
|
|
|||
Publications |
||||
Steven M. Berry, Madelyn H. Baker, Nicole J. Reardon, “Reduction potential variations in azurin through secondary coordination sphere phenylalanine incorporations” J. Inorg. Biochem. 104 (10) 1071-1078 (2010). Viehweg, Julie A.; Stamps, Sarah M.; Dertinger, Jennifer J.; Green, Robert L.; Harris, Katherine E.; Butcher, Raymond J.; Andriole, Erica J.; Poutsma, J. C.; Berry, Steven M.; Bebout, Deborah C., “A synthetic model of Hg(II) sequestration,” Dalton Transactions 39 (13) 3174-3176 (2010). Jason D. Dorweiler, Victor N. Nemykin, Amanda N. Ley, Robert D. Pike, and Steven M. Berry, “Structural and NMR Characterization of Sm(III), Eu(III), and Yb(III) Complexes of an Amide Based Polydentate Ligand Exhibiting Paramagnetic Chemical Exchange Saturation Transfer Abilities,” Inorg. Chem. 48, (19) 9365-9376 (2009). Steven M. Berry, Jonathan R. Mayers, Nicholas A. Zehm, “Models of Noncoupled Dinuclear Copper Centers in Azurin,” J. Biol. Inorg. Chem. , 14 , (1) 141-148 (2009). Madelyn H. Baker, Jason D. Dorweiler, Amanda N. Ley, Robert D. Pike, and Steven M. Berry, “Structure and emission spectra of dinuclear lanthanide(III) beta-diketonate complexes with a bridging 2,2'-bipyrimidine ligand,” Polyhedron , 28 , (1), 188-194 (2009). Deborah C. Bebout, Wei Lai, Sarah M. Stamps, Steven M. Berry, and Raymond J. Butcher, “Bis-tridentate chelates of an asymmetric ligand: X-ray structures and solution NMR characterization of divalent zinc triad metal ion complexes of N-(2-pyridylmethyl)-N-(2-(methylthio)ethyl)amine,” Polyhedron 27 , 6, 1591-1600 (2008). Deborah C. Bebout, Wei Lai, Sarah M. Stamps, Steven M. Berry, and Raymond J. Butcher, “Zinc triad metal ion complexes of NN'S ligand N-(2-pyridylmethyl)-N-(2-(methylthio)ethyl)amine,” Main Group Chemistry 6 , (3 & 4), 155-168 (2007). Lu, Y.; Berry , S. M., “Protein structure design and engineering,” Handbook of Proteins 2, 1153-1157 (2007). Garner, Dewain K.; Vaughan, Mark D.; Hwang, Hee Jung; Savelieff, Masha G.; Berry, Steven M.; Honek, John F.; Lu, Yi, “Reduction Potential Tuning of the Blue Copper Center in Pseudomonas aeruginosa Azurin by the Axial Methionine as Probed by Unnatural Amino Acids,” J. Am. Chem. Soc. 128 , 1560 8-15617 (2006). Deborah C. Bebout, Steven M. Berry, “Probing Mercury Complex Speciation with Multinuclear NMR” in Structure and Bonding: Recent Developments in Mercury Science , David Atwood Ed., Springer Science, Berlin, 120 , 81-105 (2006). Wei Lai, Steven M. Berry, Deborah C. Bebout, and Raymond J. Butcher, “Investigation of Group 12 Metal Complexes with a Tridentate SNS Ligand by X-ray Crystallography and 1 H NMR Spectroscopy” Inorg. Chem. 42 , 571-581 (2006). Hee Jung Hwang, Steven M. Berry, Mark J. Nilges and Yi Lu, “Axial Methionine in CuA Center is Much Less Influential on Reduction Potentials than in Blue Copper Center,” J. Am. Chem. Soc. 127 , 7274-7275 (2005). Yi Lu and Steven M. Berry, "Protein Structure Design and Engineering," Encyclopedia of Life Sciences, Nature Publishing Groups (2005). Steven M. Berry, Raymond J. Butcher and Deborah C. Bebout, “Solid-State and Solution-State Coordination Chemistry of the Zinc Triad with the Mixed N,S Donor Ligand Bis(2-methylpyridyl) Sulfide,” Inorg. Chem . 44 , 27-39 (2005). Martina Ralle, Steven M. Berry, Mark J. Nilges, Matt D. Gieselman, Wilfred A. van der Donk, Yi Lu and Ninian J. Blackburn, “The Selenocysteine-Substituted Blue Copper Center: Spectroscopic Investigations of Cys112SeCys Pseudomonas aeruginosa Azurin,” J. Am. Chem. Soc. 126 , 7244-7256 (2004). Steven M. Berry, Martina Ralle, Donald W. Low, Ninian J. Blackburn and Yi Lu, “Probing the Role of Axial Methionine in the Blue Copper Center of Azurin with Unnatural Amino Acids,” J. Am. Chem. Soc . 125 , 8760-8768 (2003). Steven M. Berry, Matt D. Gieselman, Mark J. Nilges, Wilfred A. van der Donk and Yi Lu, “An Engineered Azurin Variant Containing a Selenocysteine Copper Ligand,” J. Am. Chem. Soc . 124 , 2084-2085 (2002). Dmitriy Lukoyanov, Steven M. Berry, Yi Lu, William E. Antholine and Charles P. Scholes, “Role of the Coordinating Histidine in Altering the Mixed Valency of Cu A : An Electron Nuclear Double Resonance-Electron Paramagnetic Resonance Investigation,” Biophys. J . 82 , 2758-2766 (2002). Yi Lu, Steven M. Berry and Thomas D. Pfister, "Engineering Novel Metalloproteins: Design of Metal-Binding Sites into Native Protein Scaffolds" Chem. Rev . 101 , 3047-3080 (2001). L.C. Thompson and S. Berry , “Structure and emission spectrum of the o-phenanthroline adduct of tris(6-methyl-2,4-heptanedionato)europium(III),” J. Alloys Compd . 323-324 , 177-180 (2001). Steven M. Berry, Xiaotang Wang and Yi Lu, "Ligand Replacement Study at the His120 Site of Purple Cu A Azurin," J. Inorg. Biochem . 78 (Special ICBIC9 issue), 89-95 (2000). Xiaotang Wang, Steven M. Berry, Yaomin Xia and Yi Lu, "The Role of Histidine Ligands in the Structure of Purple Cu A Azurin," J. Am. Chem. Soc . 121 , 7449-7450 (1999). |
||||
@ 2018 Steven M. Berry |
||||